Differential Gene Expression Analysis Tutorial
This page outlines the ease in which differential gene expression data can be pre-processed using GEO2R. The example used is series GSE28146 from Blalock et al..
Citations
- Barrett, T., et al. (2013). NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Research, 41(D1), D991–D995.
- Blalock EM, et al. (2011). Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer's disease. J Chem Neuroanat Oct;42(2):118-26.
Step 1
Visit the link of the dataset. For this tutorial, we will use https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE28146 from Blalock et al. (2011).
Step 2
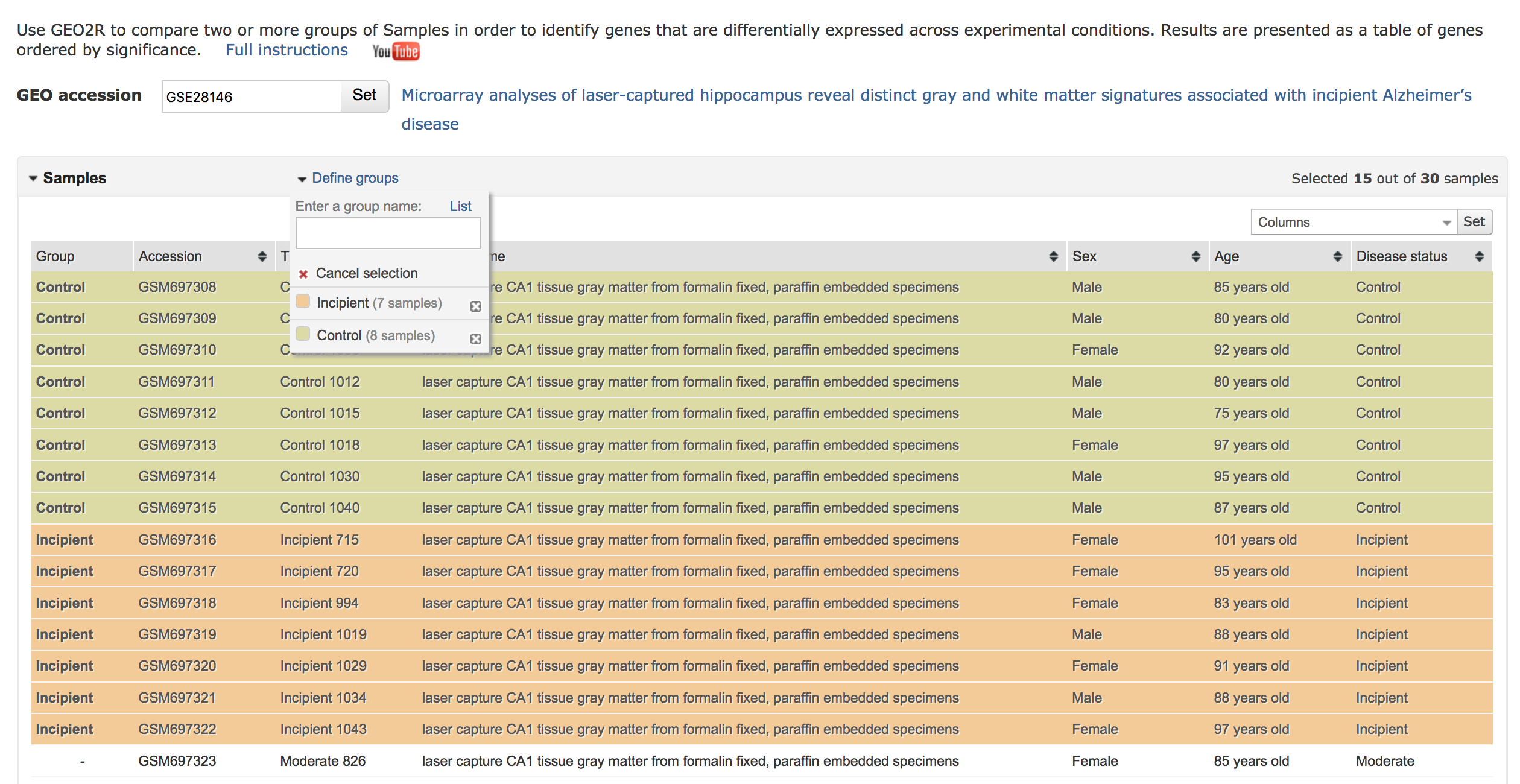
Select the corresponding groups to compare. The following steps must be repeated for each comparison. In this example, GSE28146 is analyzed 3 times to compare experiments with the following labels:
- Control vs. Incipient
- Control vs. Moderate
- Control vs. Severe
Step 3
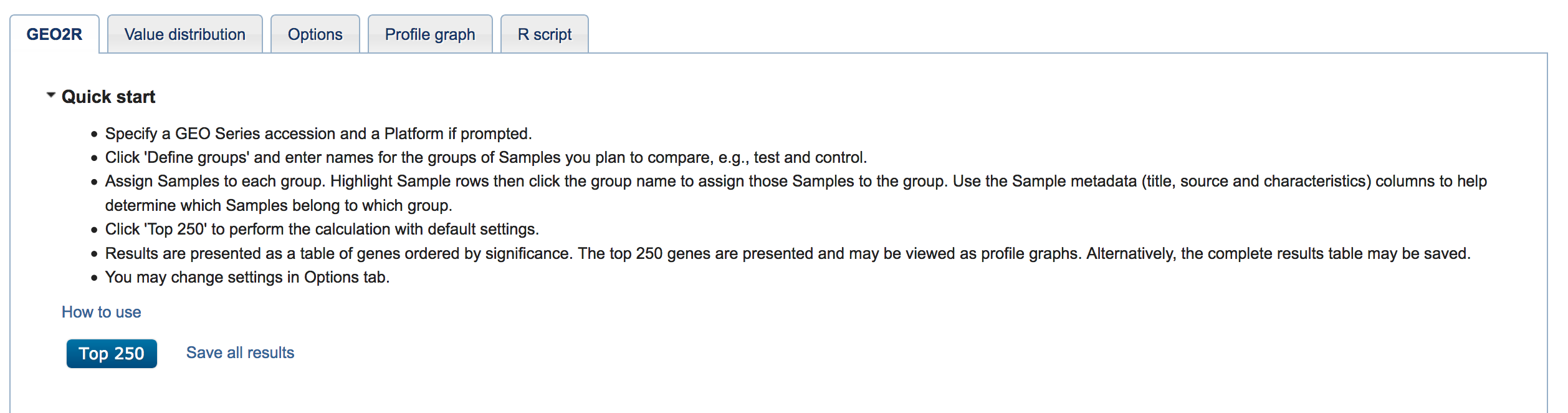
Scroll down, then click "Top 250" to perform the analysis with the default settings. If you want to change the settings, then click the "Options" tab.
Step 4
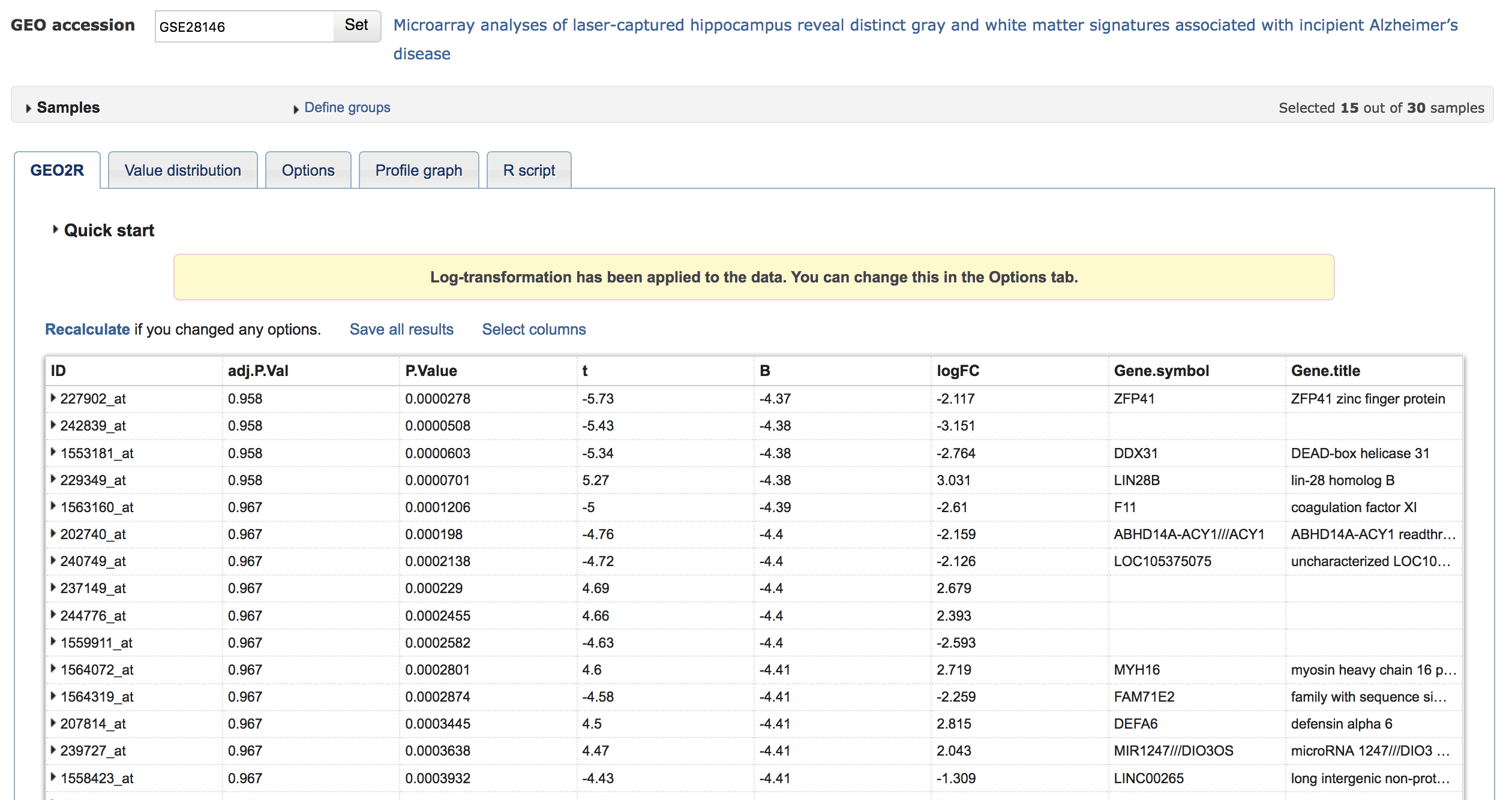
Wait (about a minute) for the analysis to run. The "Quick Start" box will be replaced with the results; after which you can can select the columns and export. You must minimally select logFC and Gene.symbol to run the heat diffusion workflow in BEL Commons. Finally, click "Save all results".
Step 5
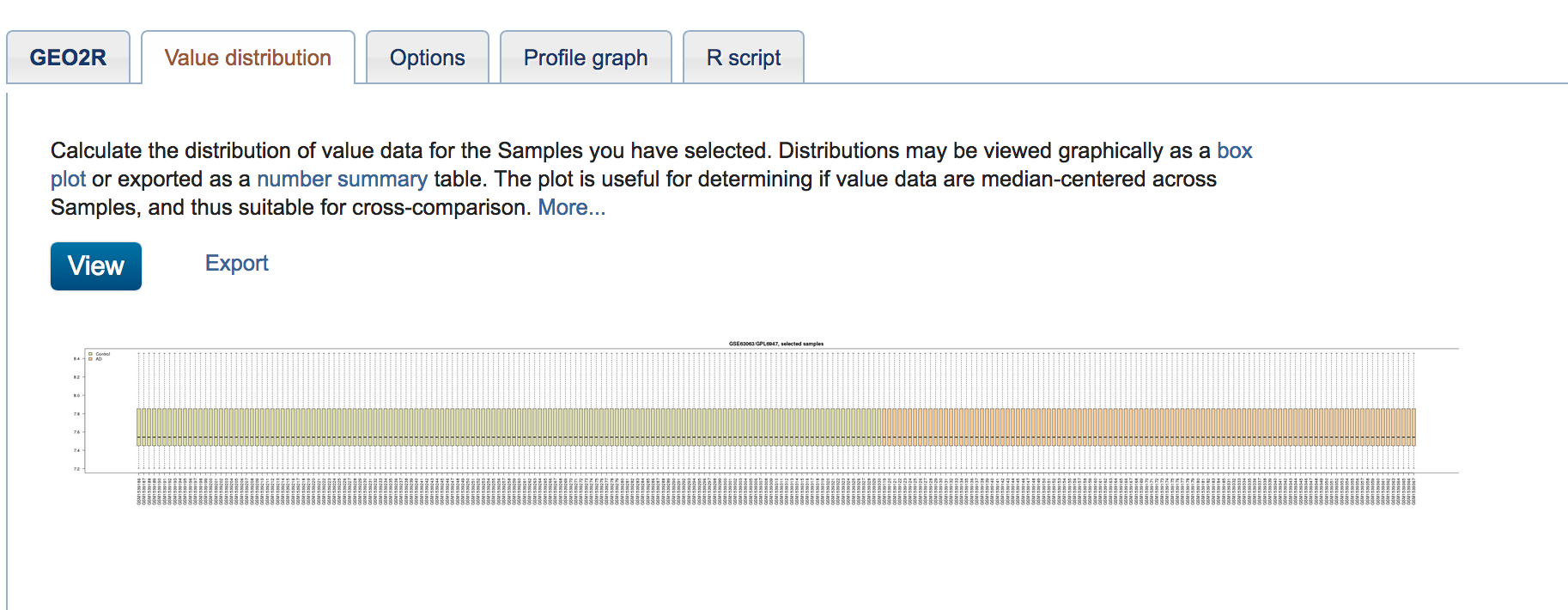
As a quality control step, click "Value Distribution" to check for outliers within the groups you have defined.